Purpose
In this experiment, we were observing bacterial growth and transforming the bacteria. The process allowed us to further understand how transformation occurs, what happens biologically to the bacteria when genes are moved, and the significance this process has on life in both prokaryotic and eukaryotic cells.
Intro
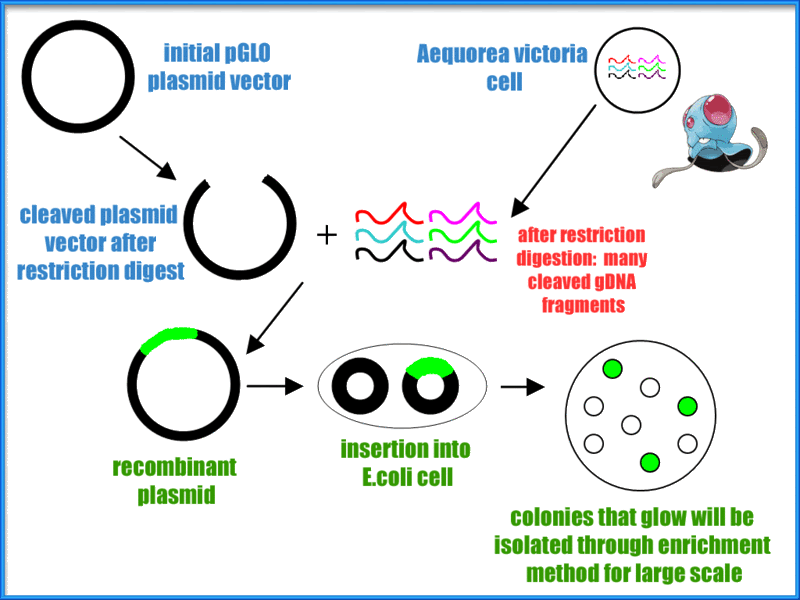
Transformation is a process used in prokaryotic as well as eukaryotic life cycles to genetically modify a cell by introducing separate DNA into the cell with the help of a plasmid. Plasmids are genetic structures in a cell that are often used in laboratories to manipulate genes. They are small, circular DNA molecules in the cytoplasm of cells that carry supplementary genes and replicate independently of the chromosome. They can even lead a cell to being antibiotic resistant. The pGLO plasmid contains genes for green florescence (GFP) as well as a gene that has resistance to the antibiotic ampicillin. Scientists such as biochemists sometimes use a method called heat shock which is where cells are put into a higher than ideal temperature of the organism. This method opens up pores in the plasma membrane due to the sudden increase in temperature, allowing plasmids to enter the bacterial cell.
Methods
Because this lab involved E.coli and making it antibiotic resistant, extra precautions, sterile equipment, and good lab techniques were used while doing this lab. No one was harmed during this genetic transformation.
Micro test tubes were labeled +pGLO and -pGLO and transformation solution was added into each tube.
All the while both test tubes were immersed in a cup of ice in order to keep them cold. Then we grabbed another sterile loop and obtained pGLO plasmid DNA where we only placed into the +pGLO and not the -pGLO. After ten minutes sitting in the cup of ice, we heat shocked the test tubes. This meant putting both tubes into a 42 degree Celsius water bath for 50 seconds then placing it back into the ice immediately afterwards. Next, LB nutrient broth was placed into both test tubes. Finally, we placed the +pGLO into the LB/amp and LB/amp/ara transformation plates, the -pGLO was placed into the LB/amp and LB control plates, and gently spread the the liquid across the plate. The only thing left to do was to put them into the incubator and wait.
The glowing E. Coli under ultraviolet light. We have a large culture here, effectively telling us that the lab was an overall success.
A different culture with even better growth than the previous one. This culture happens to include the E. Coli that were antibiotic resistant to the Ampicillin
E. Coli alone in nutrient broth
Data
Discussion
Our E. Coli pGLO transformation lab had the best results out of any lab we have done this school year. Plentiful colonies of E. Coli were produced on the control LB plate for both the wild-type and recombinant pGLO E. Coli. In terms of recombination efficiency, it was a complete success: we had one of the highest efficiencies of any lab group. Recombinant E. Coli flourished on the LB plate with ampicillin and fluoresced on the LB plate with ampicillin and arabinose as expected:
The wild-type E. Coli, as expected, did not grow on any LB plate with ampicillin. Even on these empty plates, there was very little contamination.
We contribute our success to our meticulous following of lab procedures. We used the correct amounts of materials and were not lax when it came to correct timing for the heat shock. The transfer from cold to hot to cold was immediate and precise. Our results confirmed our hypothesis perfectly: naturally, the antibiotic resistant E. Coli would grow in an ampicillin rich environment, while the wild-type would not. Overall, we are very pleased with our results.
Conclusion
By shining ultraviolet light on the petri dish and seeing E. Coli glow in the dark, we had visual proof that the plasmid had been successfully incorporated into the bacteria cells. In addition to this, only bacteria with the inserted plasmid could grow on the ampicillin culture, which also tells us that this population of E. Coli is resistant antibiotics. This lab is designed to show how genetic engineering works and the fascinating results following the simple insertion of just one new gene to the bacteria's genome. This practice is something scientists are studying on a much larger scale with the hope that humans may find additional benefits from using restriction enzymes to alter the human genome.
By shining ultraviolet light on the petri dish and seeing E. Coli glow in the dark, we had visual proof that the plasmid had been successfully incorporated into the bacteria cells. In addition to this, only bacteria with the inserted plasmid could grow on the ampicillin culture, which also tells us that this population of E. Coli is resistant antibiotics. This lab is designed to show how genetic engineering works and the fascinating results following the simple insertion of just one new gene to the bacteria's genome. This practice is something scientists are studying on a much larger scale with the hope that humans may find additional benefits from using restriction enzymes to alter the human genome.
References
http://www.cliffsnotes.com/sciences/biology/microbiology/microbial-genetics/the-bacterial-chromosome-and-plasmid
en.wikipedia.org/wiki/Heat_shock
http://www.jove.com/science-education/5059/bacterial-transformation-the-heat-shock-method
en.wikipedia.org/wiki/Heat_shock
http://www.jove.com/science-education/5059/bacterial-transformation-the-heat-shock-method










No comments:
Post a Comment